Single cell RNAseq data
Finding gene expression data on VFB
Finding cell type clusters
Cell types with available scRNAseq data can be identified using the ‘Has scRNAseq data filter’ when searching.
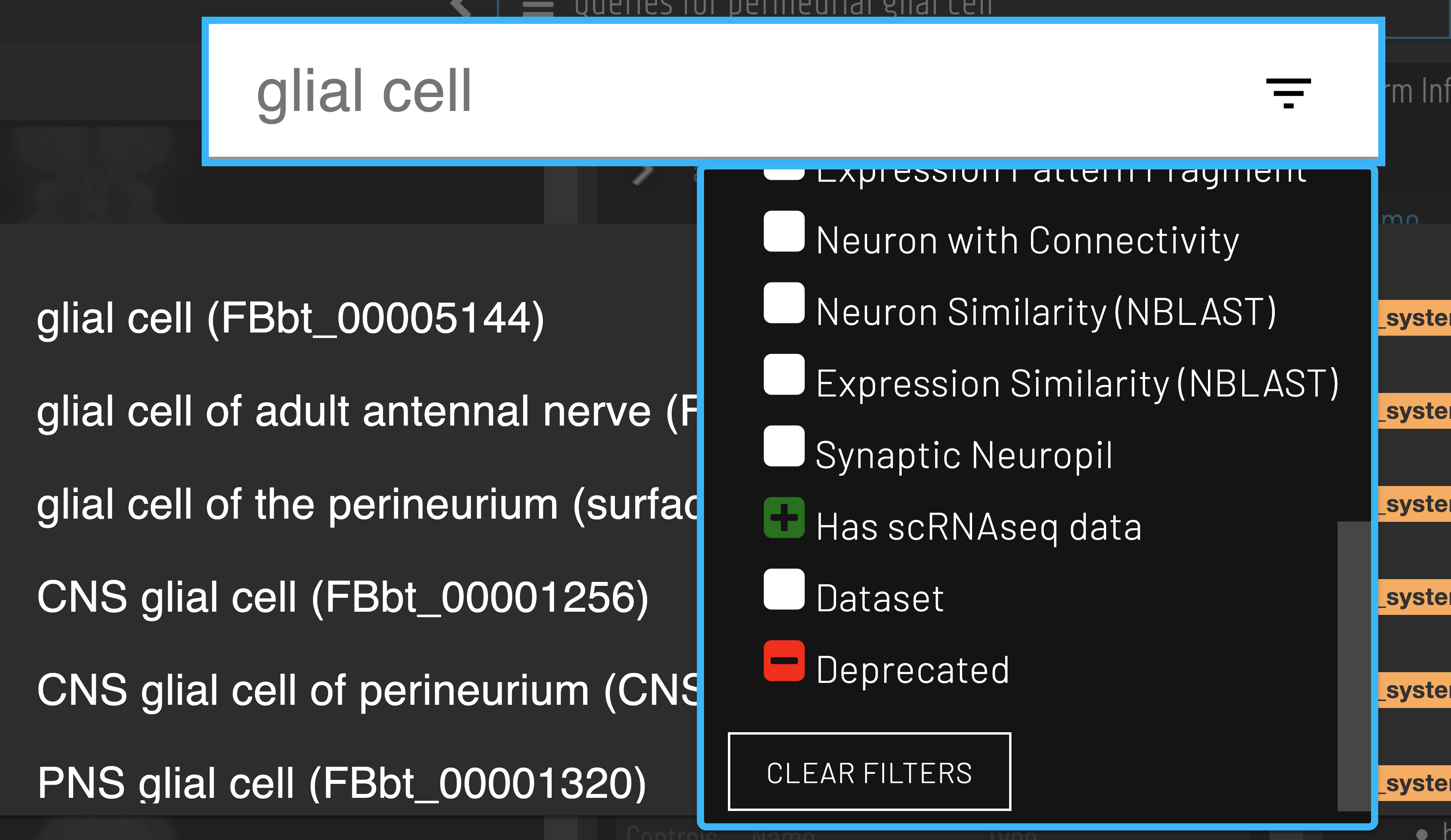
Clusters for a cell type of interest can be found via the ‘Single cell transcriptomics data for…’ query.

Gene expression data
After selecting a cluster, gene expression can be retrieved via the ‘Genes expressed in…’ query.
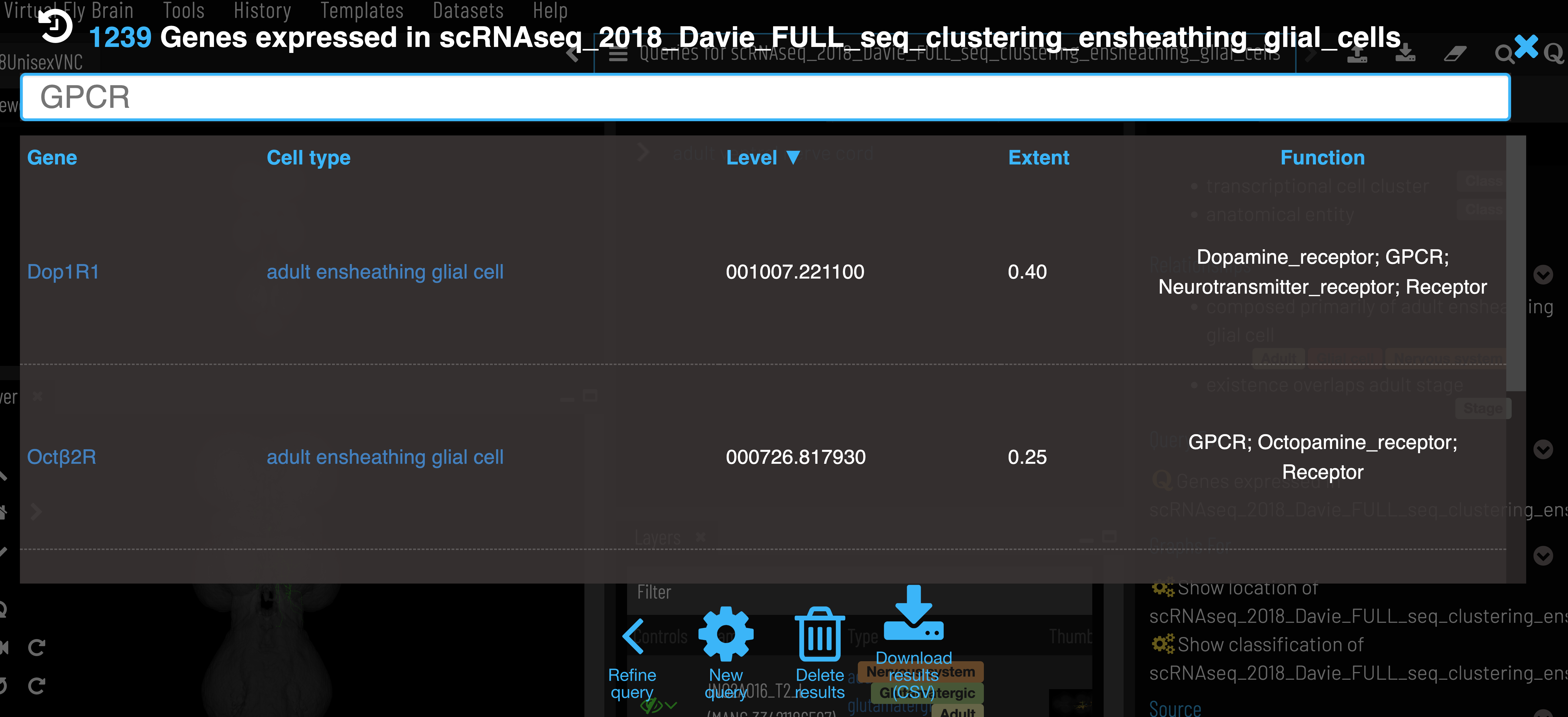
Expression level is the mean counts per million reads of all cells in the cell type cluster that express the given gene. Expression extent is the proportion of cells within the cluster that express the given gene. VFB only shows genes that are expressed in at least 20% of cells in the cluster (Extent > 0.2).
Gene semantic tags
We add semantic tags to genes to allow quick searching and filtering of results (‘Function’ column of gene expression data). These tags are based on FlyBase Gene Group membership and GO annotations, which are all sourced from FlyBase. A full list of gene tags with their associated Gene group and GO terms can be found here.
API
Data can also be retrieved using VFB_connect.
Data sources and processing
- Raw scRNAseq data is ingested by the EBI Single Cell Expression Atlas (SCEA). Where possible, cells are linked to cell types from the Drosophila Anatomy Ontology based on author annotations. Data is reprocessed, filtered and reclustered.
- FlyBase takes data from EBI, keeping only cells that are linked to ontology terms and generating summary expression data for each cell type (cluster).
- VFB takes control/wild type expression data from FlyBase for datasets where at least one nervous system cell type is present and filter out any genes expressed in less than 20% of cells per cluster.
Database schema
A simplified schema showing how scRNAseq data is stored in the VFB Neo4j database is shown below:
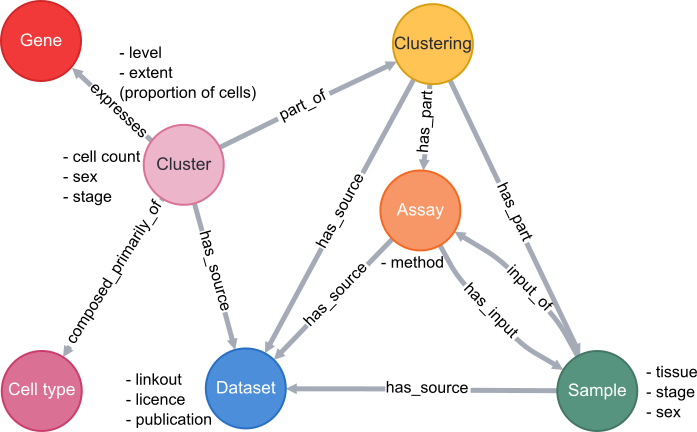
Cell types are classes from the Drosophila Anatomy Ontology and are also linked to several other types of data in VFB, such as images, connectomics and driver expression information.
Feedback
Was this page helpful?
Glad to hear it! Please tell us how we can improve.
Sorry to hear that. Please tell us how we can improve.